Automated Nucleic Acid Isolation from seed sample
Simplifying chickpea seed sample preparation: An integrated solution using the Bead Ruptor™ 96 Bead Mill Homogenizer and chemagic™ 360 Nucleic Acid Extractor
Seeds, nuts, and other plant matrices are inherently resistant to fast and consistent sample preparation for downstream analysis. Large seeds, specifically, have unique challenges that typically require time consuming pre-processing steps and/or intensive cleaning procedures to reduce cross carryover between samples. These traditional methods can hinder high throughput workflows. Additionally, standard protocols for DNA or other analyte purification methods require powdered matrices prior to mixing with an aqueous buffer that is compatible with chosen downstream extraction. This process can be labor intensive, inexact, and time consuming.
In this application note, we discuss a sample preparation method for large seeds, using the Bead Ruptor™ 96 Bead Mill Homogenizer, that allows samples to be processed in both buffered and dry conditions which results in high quality and quantity DNA. Therefore, this method can prepare samples with a high degree of throughput and is compatible with automated DNA extraction using the chemagic™ 360 Nucleic Acid Extractor.
Automated nucleic acid isolation from seed sample
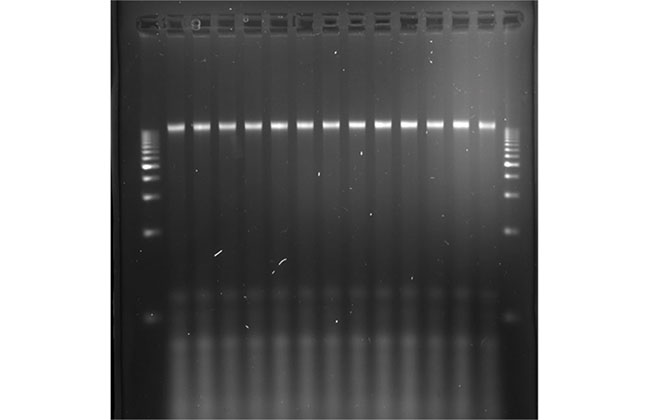
High quality and quantity DNA was extracted from chickpea seeds
2 % agarose gel from extracted DNA using dry grinding as a sample processing step.
Procedure
Exactly 6 chickpea seeds were chosen as a representative sample size for nucleic acid extraction. Each sample of chickpea seeds was loaded into 50 mL conical tubes. Two processing parameters (dry grinding, and liquid homogenization) were evaluated for DNA extractions and are detailed in the application note below, along with downstream methodologies.
For more information, please download the application note about automated nucleic acid extraction from chickpea seed samples.
For research use only. Not for use in diagnostic procedures.
